Daniel W. Lin, MD, presented “Genomic Markers and DNA Sequencing Testing” during the 19th Annual Future Directions in Urology Symposium on August 11, 2018, in Colorado Springs, Colorado.
How to cite: Lin, Daniel W. “Genomic Markers and DNA Sequencing Testing” August 12, 2018. Accessed [date today]. https://grandroundsinurology.com/genomic-markers-and-dna-sequencing-testing/
Genomic Markers and DNA Sequencing Testing – Summary
Daniel W. Lin, MD, reviews novel and emerging biomarkers across the spectrum of prostate cancer. He then explains an emerging model of prostate cancer treatment with genomic markers and DNA sequencing, discussing newly-identified precision targets and their therapeutic decision-making utility.
Learning from Advances in Biomarkers for Breast Cancer
In the setting of breast cancer, estrogen receptor/ progesterone receptor ER/PR positive and human epidermal growth factor receptor 2 (HER2) biomarkers, as well as the Oncotype DX 21 gene assay, have been in use for many years. Furthermore, the recent TAILORx study, the Oncotype DX proved to be an accurate tool for determining which women would not benefit from chemotherapy.
Dr. Lin argues that these advancements biomarkers for breast cancer put the lack of research and developments in biomarkers for prostate cancer into sharp relief. For example, there are no National Comprehensive Cancer Network (NCCN) guidelines regarding biomarkers for prostate cancer.
Important Biomarker Criteria
In order to adopt a biomarker into clinical practice, the marker must have feasibility and a well-founded biologic rationale. The marker must also have appropriate application and validation.
It must add to established predictive models based on readily available clinical and pathological data. In other words, a marker must provide further information that PSA, Gleason stage, biopsy, age, race, and family do not already provide.
Finally, the biomarker must have utility in physicians’ pre- and post-therapy decision making, and impact disease-specific outcomes.
Overview of Current Biomarkers for Prostate Cancer
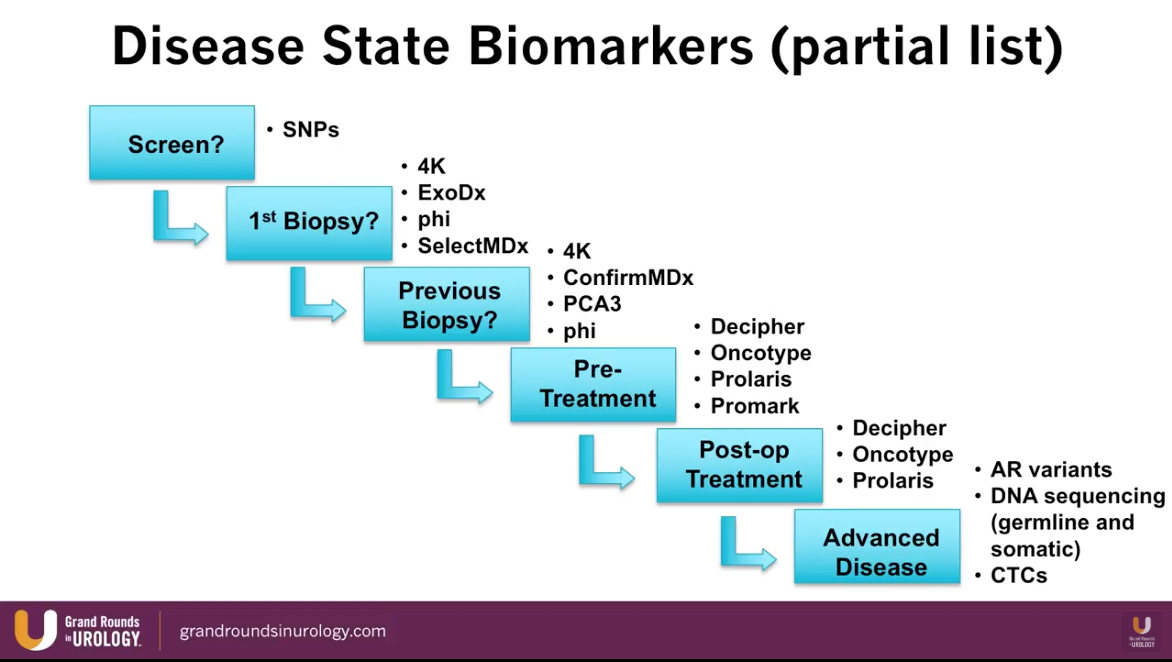
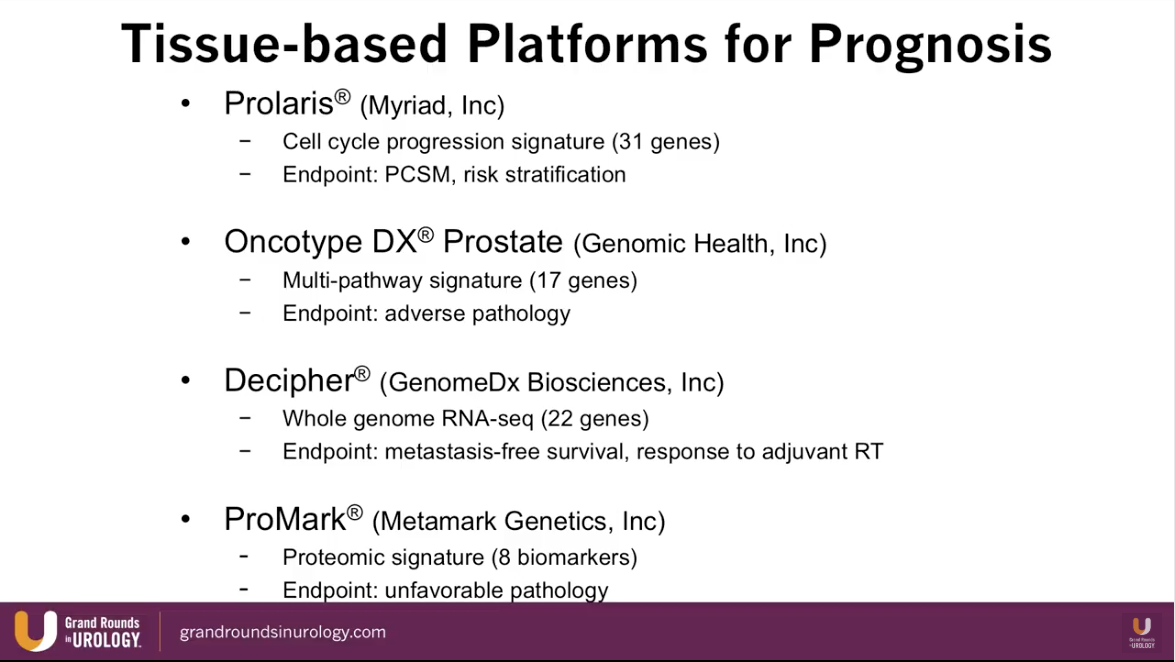
Dr. Lin presents an abbreviated list of currently available biomarkers for prostate cancer and maps each one into their time of utilization according to stage of prostate cancer treatment, as illustrated to the left.
Specifically, the SelectMDx and ConfirmMDx assess patients in the pre-diagnostic setting. The Select MDx is a urinary biomarker currently available in Europe. A study by Van Neste et al. showed that Select MDx had a negative predictive value (NPV) of 93%-94% for high-grade prostate cancer. A separate study by Van Neste et al. showed that ConfirmMDx had a NPV of 96% for high-grade prostate cancer. Dr. Lin notes that NPV is the most important performance measure when evaluating the quality of a biomarker.
Furthermore, a phase II NRG Oncology trial is comparing salvage radiation (SRT) plus apalutamide against SRT alone in men with post-prostatectomy PSA recurrences. This study uses a biomarker to identify patients with a luminal B subtype of prostate cancer, therefore identifying which patients may benefit from the addition of apalutamide. Dr. Lin believes it is important to incorporate biomarkers into trial designs, as well as using biomarkers to stratify which patients to include in a trial.
Emerging Model of Cancer Treatment
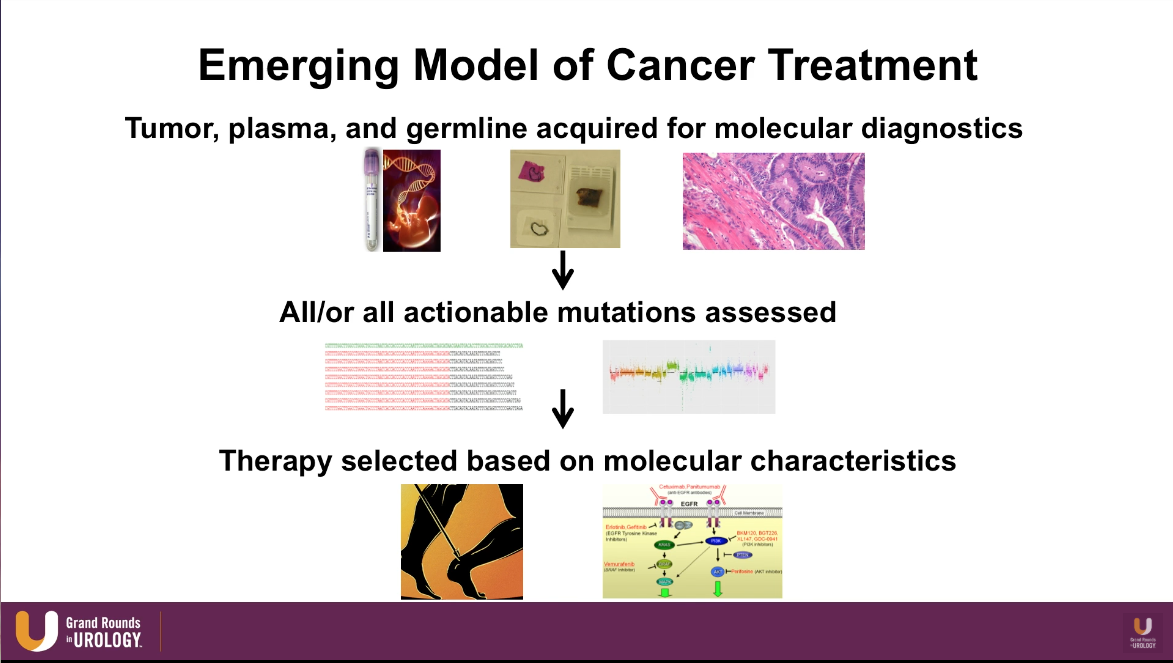
The emerging model of cancer treatment heavily incorporates genomic markers and DNA sequencing. This model begins with acquiring tumor, plasma, and germline and assessing all the actionable mutations from those samples. Then, physicians can select therapy and pathways to target based on those molecular characteristics.
Germline DNA Repair Mutations and Emerging Precision Targets
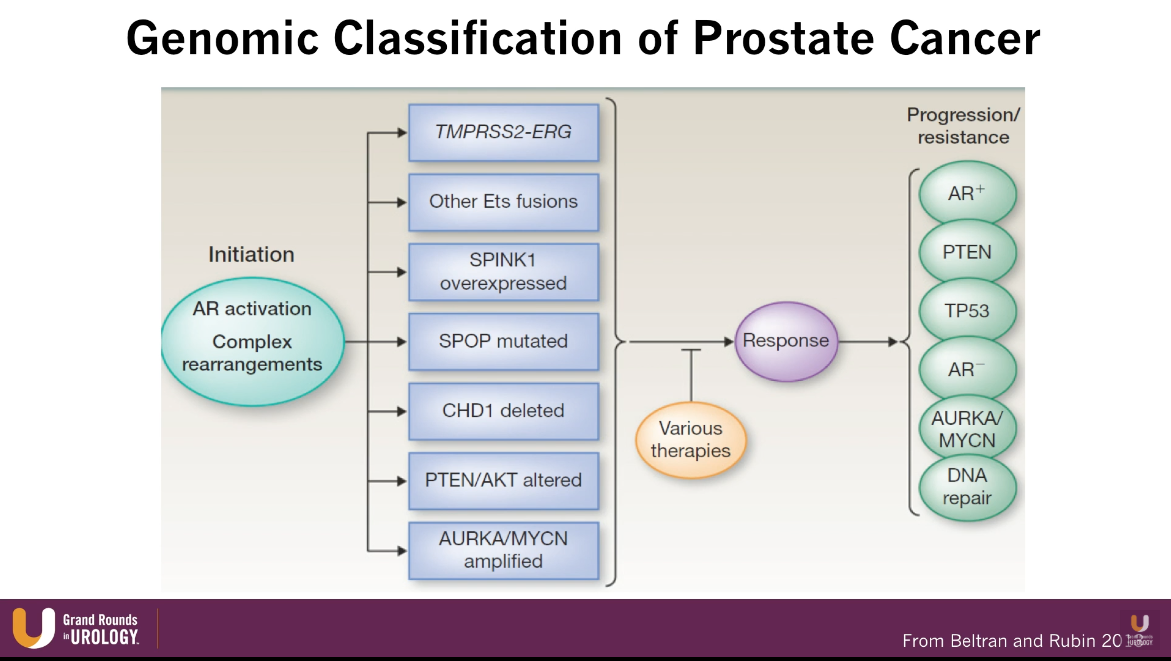
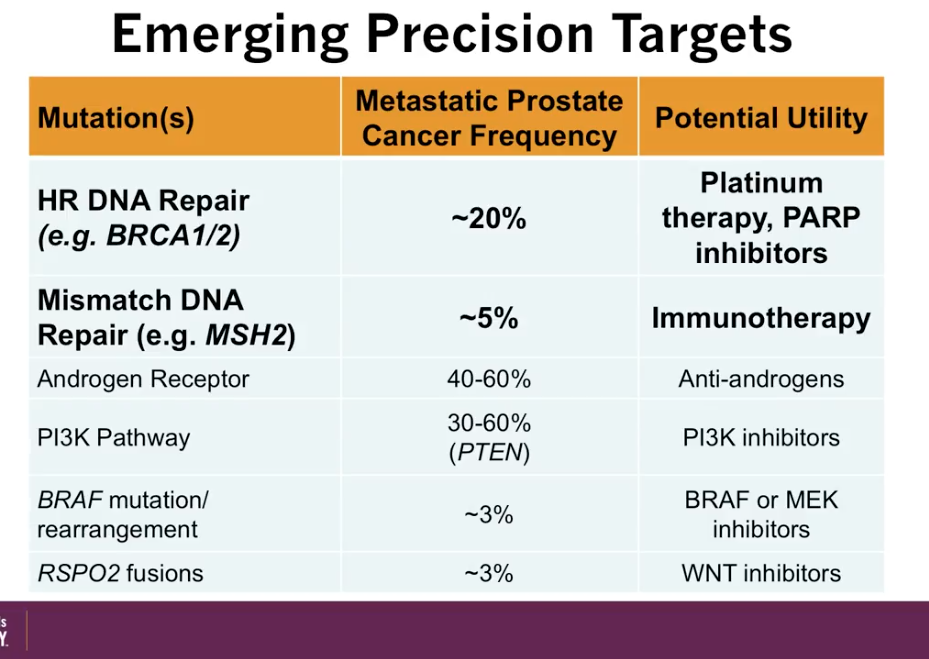
According to an article by Pritchard et al., out of all patients with metastatic prostate cancer, about 12% have a germline DNA repair mutations, such as BRCA 1 and 2. These patients are particularly sensitive to platinum therapies. Moreover, patients with mismatch DNA repair mutations, such as MSH2, are particularly sensitive to immunotherapy. The figure to the left illustrates further potential utilities correlating with specific mutations.
Finally, there are multiple ongoing phase II and III trials relevant to DNA repair defects in mCRPC. Most notably, the TRITON3 trial is comparing rucaparib against abiraterone, enzalutamide, or docetaxel in mCRPC patients with BRCA 1 or 2 or Ataxia-Telangiesctasia (ATM) mutations. Similarly, the PROfound trial is comparing olaparib against enzalutamide or abiraterone in mCRPC patients with a somatic homologous recombination deficient (HRD) mutation. Hopefully, these trials will provide answers as to the benefit of using these targets in therapeutic decision-making.
ABOUT THE AUTHOR
Dr. Lin is the Pritt Family Endowed Chair for Prostate Cancer Research and Professor and Chief of Urologic Oncology in the Department of Urology at the University of Washington School of Medicine in Seattle. He is also a urologist at the University of Washington Medical Center and the Seattle Cancer Care Alliance. He received his medical degree at Vanderbilt University Medical Center in Nashville, Tennessee, pursued a Fellowship in Urologic Oncology at Memorial Sloan-Kettering Cancer Center in New York City, and completed his internship and residency at the University of Washington School of Medicine. He serves on the National Comprehensive Cancer Network Guideline Panel for Renal and Testis Tumors, the Society of Urologic Oncology Executive Board, and the AUA Guideline Committees for Advanced Prostate Cancer and Renal Mass Follow-Up. Dr. Lin’s research interests include prostate chemoprevention and carcinogenesis, and his clinical research efforts are in active surveillance of prostate cancer and management of high-risk prostate cancer.